使用机器学习进行疾病预测
本文旨在实现一个强大的机器学习模型,该模型可以根据人类的症状有效地预测人类的疾病。让我们看看如何解决这个机器学习问题:
方法:
- 收集数据:数据准备是任何机器学习问题的首要步骤。我们将使用 Kaggle 的数据集来解决这个问题。该数据集由两个 CSV 文件组成,一个用于训练,一个用于测试。数据集中共有 133 列,其中 132 列代表症状,最后一列是预后。
- 清理数据:清理是机器学习项目中最重要的一步。我们数据的质量决定了我们机器学习模型的质量。因此,在将数据提供给模型进行训练之前,始终需要清理数据。在我们的数据集中,所有列都是数字,目标列即预测是字符串类型,并使用标签编码器编码为数字形式。
- 模型构建:收集和清理数据后,数据准备就绪,可用于训练机器学习模型。我们将使用这些清理过的数据来训练支持向量分类器、朴素贝叶斯分类器和随机森林分类器。我们将使用混淆矩阵来确定模型的质量。
- 推理:训练三个模型后,我们将通过结合所有三个模型的预测来预测输入症状的疾病。这使我们的整体预测更加稳健和准确。
最后,我们将定义一个函数,该函数以逗号分隔的症状作为输入,使用经过训练的模型根据症状预测疾病,并以 JSON 格式返回预测结果。
实施工作流程:

确保下载了培训和测试,并将 train.csv、test.csv 放在数据集文件夹中。打开 jupyter notebook 并单独运行代码以更好地理解。
Python3
# Importing libraries
import numpy as np
import pandas as pd
from scipy.stats import mode
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.svm import SVC
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score, confusion_matrix
%matplotlib inlinePython3
# Reading the train.csv by removing the
# last column since it's an empty column
DATA_PATH = "dataset/Training.csv"
data = pd.read_csv(DATA_PATH).dropna(axis = 1)
# Checking whether the dataset is balanced or not
disease_counts = data["prognosis"].value_counts()
temp_df = pd.DataFrame({
"Disease": disease_counts.index,
"Counts": disease_counts.values
})
plt.figure(figsize = (18,8))
sns.barplot(x = "Disease", y = "Counts", data = temp_df)
plt.xticks(rotation=90)
plt.show()Python3
# Encoding the target value into numerical
# value using LabelEncoder
encoder = LabelEncoder()
data["prognosis"] = encoder.fit_transform(data["prognosis"])Python3
X = data.iloc[:,:-1]
y = data.iloc[:, -1]
X_train, X_test, y_train, y_test =train_test_split(
X, y, test_size = 0.2, random_state = 24)
print(f"Train: {X_train.shape}, {y_train.shape}")
print(f"Test: {X_test.shape}, {y_test.shape}")Python3
# Defining scoring metric for k-fold cross validation
def cv_scoring(estimator, X, y):
return accuracy_score(y, estimator.predict(X))
# Initializing Models
models = {
"SVC":SVC(),
"Gaussian NB":GaussianNB(),
"Random Forest":RandomForestClassifier(random_state=18)
}
# Producing cross validation score for the models
for model_name in models:
model = models[model_name]
scores = cross_val_score(model, X, y, cv = 10,
n_jobs = -1,
scoring = cv_scoring)
print("=="*30)
print(model_name)
print(f"Scores: {scores}")
print(f"Mean Score: {np.mean(scores)}")Python3
# Training and testing SVM Classifier
svm_model = SVC()
svm_model.fit(X_train, y_train)
preds = svm_model.predict(X_test)
print(f"Accuracy on train data by SVM Classifier\
: {accuracy_score(y_train, svm_model.predict(X_train))*100}")
print(f"Accuracy on test data by SVM Classifier\
: {accuracy_score(y_test, preds)*100}")
cf_matrix = confusion_matrix(y_test, preds)
plt.figure(figsize=(12,8))
sns.heatmap(cf_matrix, annot=True)
plt.title("Confusion Matrix for SVM Classifier on Test Data")
plt.show()
# Training and testing Naive Bayes Classifier
nb_model = GaussianNB()
nb_model.fit(X_train, y_train)
preds = nb_model.predict(X_test)
print(f"Accuracy on train data by Naive Bayes Classifier\
: {accuracy_score(y_train, nb_model.predict(X_train))*100}")
print(f"Accuracy on test data by Naive Bayes Classifier\
: {accuracy_score(y_test, preds)*100}")
cf_matrix = confusion_matrix(y_test, preds)
plt.figure(figsize=(12,8))
sns.heatmap(cf_matrix, annot=True)
plt.title("Confusion Matrix for Naive Bayes Classifier on Test Data")
plt.show()
# Training and testing Random Forest Classifier
rf_model = RandomForestClassifier(random_state=18)
rf_model.fit(X_train, y_train)
preds = rf_model.predict(X_test)
print(f"Accuracy on train data by Random Forest Classifier\
: {accuracy_score(y_train, rf_model.predict(X_train))*100}")
print(f"Accuracy on test data by Random Forest Classifier\
: {accuracy_score(y_test, preds)*100}")
cf_matrix = confusion_matrix(y_test, preds)
plt.figure(figsize=(12,8))
sns.heatmap(cf_matrix, annot=True)
plt.title("Confusion Matrix for Random Forest Classifier on Test Data")
plt.show()Python3
# Training the models on whole data
final_svm_model = SVC()
final_nb_model = GaussianNB()
final_rf_model = RandomForestClassifier(random_state=18)
final_svm_model.fit(X, y)
final_nb_model.fit(X, y)
final_rf_model.fit(X, y)
# Reading the test data
test_data = pd.read_csv("./dataset/Testing.csv").dropna(axis=1)
test_X = test_data.iloc[:, :-1]
test_Y = encoder.transform(test_data.iloc[:, -1])
# Making prediction by take mode of predictions
# made by all the classifiers
svm_preds = final_svm_model.predict(test_X)
nb_preds = final_nb_model.predict(test_X)
rf_preds = final_rf_model.predict(test_X)
final_preds = [mode([i,j,k])[0][0] for i,j,
k in zip(svm_preds, nb_preds, rf_preds)]
print(f"Accuracy on Test dataset by the combined model\
: {accuracy_score(test_Y, final_preds)*100}")
cf_matrix = confusion_matrix(test_Y, final_preds)
plt.figure(figsize=(12,8))
sns.heatmap(cf_matrix, annot = True)
plt.title("Confusion Matrix for Combined Model on Test Dataset")
plt.show()Python3
symptoms = X.columns.values
# Creating a symptom index dictionary to encode the
# input symptoms into numerical form
symptom_index = {}
for index, value in enumerate(symptoms):
symptom = " ".join([i.capitalize() for i in value.split("_")])
symptom_index[symptom] = index
data_dict = {
"symptom_index":symptom_index,
"predictions_classes":encoder.classes_
}
# Defining the Function
# Input: string containing symptoms separated by commmas
# Output: Generated predictions by models
def predictDisease(symptoms):
symptoms = symptoms.split(",")
# creating input data for the models
input_data = [0] * len(data_dict["symptom_index"])
for symptom in symptoms:
index = data_dict["symptom_index"][symptom]
input_data[index] = 1
# reshaping the input data and converting it
# into suitable format for model predictions
input_data = np.array(input_data).reshape(1,-1)
# generating individual outputs
rf_prediction = data_dict["predictions_classes"][final_rf_model.predict(input_data)[0]]
nb_prediction = data_dict["predictions_classes"][final_nb_model.predict(input_data)[0]]
svm_prediction = data_dict["predictions_classes"][final_svm_model.predict(input_data)[0]]
# making final prediction by taking mode of all predictions
final_prediction = mode([rf_prediction, nb_prediction, svm_prediction])[0][0]
predictions = {
"rf_model_prediction": rf_prediction,
"naive_bayes_prediction": nb_prediction,
"svm_model_prediction": nb_prediction,
"final_prediction":final_prediction
}
return predictions
# Testing the function
print(predictDisease("Itching,Skin Rash,Nodal Skin Eruptions,Dischromic Patches"))读取数据集
首先,我们将使用 pandas 库从文件夹中加载数据集。在读取数据集时,我们将删除空列。这个数据集是一个干净的数据集,没有空值,所有的特征都由 0 和 1 组成。每当我们解决分类任务时,有必要检查我们的目标列是否平衡。我们将使用条形图来检查数据集是否平衡。
蟒蛇3
# Reading the train.csv by removing the
# last column since it's an empty column
DATA_PATH = "dataset/Training.csv"
data = pd.read_csv(DATA_PATH).dropna(axis = 1)
# Checking whether the dataset is balanced or not
disease_counts = data["prognosis"].value_counts()
temp_df = pd.DataFrame({
"Disease": disease_counts.index,
"Counts": disease_counts.values
})
plt.figure(figsize = (18,8))
sns.barplot(x = "Disease", y = "Counts", data = temp_df)
plt.xticks(rotation=90)
plt.show()
输出:
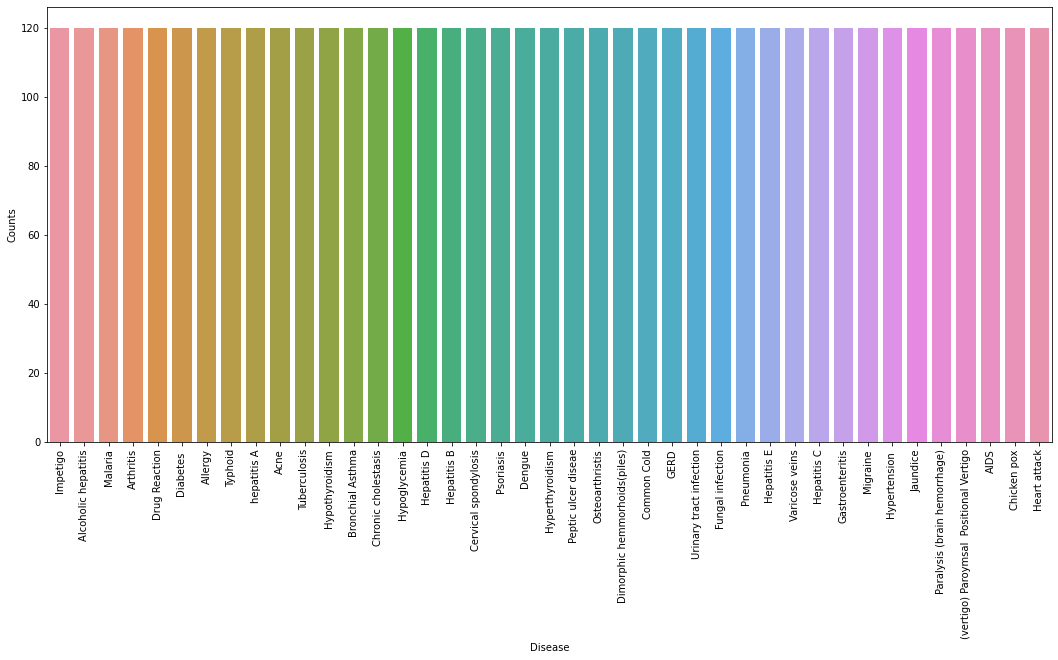
从上图中,我们可以观察到数据集是一个平衡数据集,即每种疾病正好有 120 个样本,不需要进一步的平衡。我们可以注意到我们的目标列,即预测列是对象数据类型,这种格式不适合训练机器学习模型。因此,我们将使用标签编码器将预测列转换为数值数据类型。标签编码器通过为标签分配唯一索引将标签转换为数字形式。如果标签总数为 n,则分配给每个标签的编号将在 0 到 n-1 之间。
蟒蛇3
# Encoding the target value into numerical
# value using LabelEncoder
encoder = LabelEncoder()
data["prognosis"] = encoder.fit_transform(data["prognosis"])
拆分数据以训练和测试模型
现在我们已经通过删除 Null 值并将标签转换为数字格式来清理我们的数据,是时候拆分数据来训练和测试模型了。我们将数据拆分为 80:20 格式,即 80% 的数据集将用于训练模型,20% 的数据将用于评估模型的性能。
蟒蛇3
X = data.iloc[:,:-1]
y = data.iloc[:, -1]
X_train, X_test, y_train, y_test =train_test_split(
X, y, test_size = 0.2, random_state = 24)
print(f"Train: {X_train.shape}, {y_train.shape}")
print(f"Test: {X_test.shape}, {y_test.shape}")
输出:
Train: (3936, 132), (3936,)
Test: (984, 132), (984,)建筑模型
拆分数据后,我们现在将处理建模部分。我们将使用 K-Fold 交叉验证来评估机器学习模型。我们将使用支持向量分类器、高斯朴素贝叶斯分类器和随机森林分类器进行交叉验证。在进入实现部分之前,让我们熟悉 k 折交叉验证和机器学习模型。
- K-Fold Cross-Validation: K-Fold 交叉验证是一种交叉验证技术,其中将整个数据集拆分为 k 个子集,也称为折叠,然后在 k- 1 个子集,剩下的一个子集用于评估模型性能。
- 支持向量分类器:支持向量分类器是一个判别式分类器,即当给定一个标记的训练数据时,该算法试图找到一个最佳超平面,将样本准确地分成超空间中的不同类别。
- Gaussian Naive Bayes Classifier:它是一种概率机器学习算法,内部使用贝叶斯定理对数据点进行分类。
- 随机森林分类器:随机森林是一种基于集成学习的监督机器学习分类算法,内部使用多个决策树进行分类。在随机森林分类器中,所有内部决策树都是弱学习器,这些弱决策树的输出组合在一起,即所有预测的模式作为最终预测。
使用 K 折交叉验证进行模型选择
蟒蛇3
# Defining scoring metric for k-fold cross validation
def cv_scoring(estimator, X, y):
return accuracy_score(y, estimator.predict(X))
# Initializing Models
models = {
"SVC":SVC(),
"Gaussian NB":GaussianNB(),
"Random Forest":RandomForestClassifier(random_state=18)
}
# Producing cross validation score for the models
for model_name in models:
model = models[model_name]
scores = cross_val_score(model, X, y, cv = 10,
n_jobs = -1,
scoring = cv_scoring)
print("=="*30)
print(model_name)
print(f"Scores: {scores}")
print(f"Mean Score: {np.mean(scores)}")
输出:
============================================================
SVC
Scores: [1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
Mean Score: 1.0
============================================================
Gaussian NB
Scores: [1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
Mean Score: 1.0
============================================================
Random Forest
Scores: [1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
Mean Score: 1.0
从上面的输出中,我们可以注意到我们所有的机器学习算法都表现得非常好,k 折交叉验证后的平均分数也非常高。为了建立一个健壮的模型,我们可以结合即采用所有三个模型的预测模式,这样即使一个模型做出错误的预测,而另外两个做出正确的预测,那么最终输出将是正确的。这种方法将帮助我们在完全看不见的数据上保持更准确的预测。在下面的代码中,我们将在训练数据上训练所有三个模型,使用混淆矩阵检查模型的质量,然后结合所有三个模型的预测。
通过组合所有模型构建稳健的分类器:
蟒蛇3
# Training and testing SVM Classifier
svm_model = SVC()
svm_model.fit(X_train, y_train)
preds = svm_model.predict(X_test)
print(f"Accuracy on train data by SVM Classifier\
: {accuracy_score(y_train, svm_model.predict(X_train))*100}")
print(f"Accuracy on test data by SVM Classifier\
: {accuracy_score(y_test, preds)*100}")
cf_matrix = confusion_matrix(y_test, preds)
plt.figure(figsize=(12,8))
sns.heatmap(cf_matrix, annot=True)
plt.title("Confusion Matrix for SVM Classifier on Test Data")
plt.show()
# Training and testing Naive Bayes Classifier
nb_model = GaussianNB()
nb_model.fit(X_train, y_train)
preds = nb_model.predict(X_test)
print(f"Accuracy on train data by Naive Bayes Classifier\
: {accuracy_score(y_train, nb_model.predict(X_train))*100}")
print(f"Accuracy on test data by Naive Bayes Classifier\
: {accuracy_score(y_test, preds)*100}")
cf_matrix = confusion_matrix(y_test, preds)
plt.figure(figsize=(12,8))
sns.heatmap(cf_matrix, annot=True)
plt.title("Confusion Matrix for Naive Bayes Classifier on Test Data")
plt.show()
# Training and testing Random Forest Classifier
rf_model = RandomForestClassifier(random_state=18)
rf_model.fit(X_train, y_train)
preds = rf_model.predict(X_test)
print(f"Accuracy on train data by Random Forest Classifier\
: {accuracy_score(y_train, rf_model.predict(X_train))*100}")
print(f"Accuracy on test data by Random Forest Classifier\
: {accuracy_score(y_test, preds)*100}")
cf_matrix = confusion_matrix(y_test, preds)
plt.figure(figsize=(12,8))
sns.heatmap(cf_matrix, annot=True)
plt.title("Confusion Matrix for Random Forest Classifier on Test Data")
plt.show()
输出:
Accuracy on train data by SVM Classifier: 100.0
Accuracy on test data by SVM Classifier: 100.0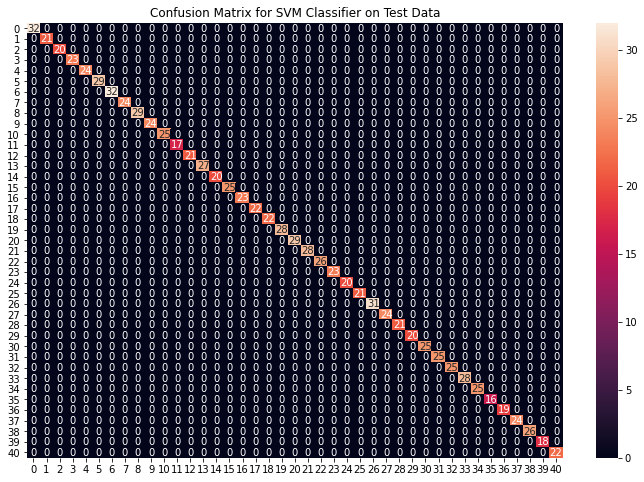
Accuracy on train data by Naive Bayes Classifier: 100.0
Accuracy on test data by Naive Bayes Classifier: 100.0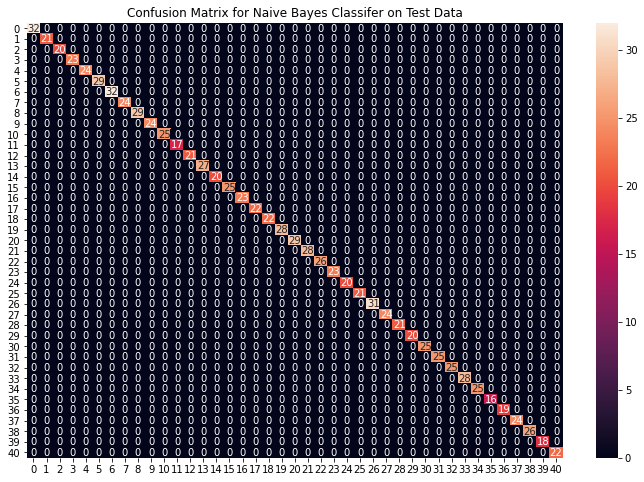
Accuracy on train data by Random Forest Classifier: 100.0
Accuracy on test data by Random Forest Classifier: 100.0
从上面的混淆矩阵中,我们可以看到模型在看不见的数据上表现得非常好。现在我们将在我们下载的数据集中存在的整个训练数据上训练模型,然后在数据集中存在的测试数据上测试我们的组合模型。
在整个数据上拟合模型并在测试数据集上进行验证:
蟒蛇3
# Training the models on whole data
final_svm_model = SVC()
final_nb_model = GaussianNB()
final_rf_model = RandomForestClassifier(random_state=18)
final_svm_model.fit(X, y)
final_nb_model.fit(X, y)
final_rf_model.fit(X, y)
# Reading the test data
test_data = pd.read_csv("./dataset/Testing.csv").dropna(axis=1)
test_X = test_data.iloc[:, :-1]
test_Y = encoder.transform(test_data.iloc[:, -1])
# Making prediction by take mode of predictions
# made by all the classifiers
svm_preds = final_svm_model.predict(test_X)
nb_preds = final_nb_model.predict(test_X)
rf_preds = final_rf_model.predict(test_X)
final_preds = [mode([i,j,k])[0][0] for i,j,
k in zip(svm_preds, nb_preds, rf_preds)]
print(f"Accuracy on Test dataset by the combined model\
: {accuracy_score(test_Y, final_preds)*100}")
cf_matrix = confusion_matrix(test_Y, final_preds)
plt.figure(figsize=(12,8))
sns.heatmap(cf_matrix, annot = True)
plt.title("Confusion Matrix for Combined Model on Test Dataset")
plt.show()
输出:
Accuracy on Test dataset by the combined model: 100.0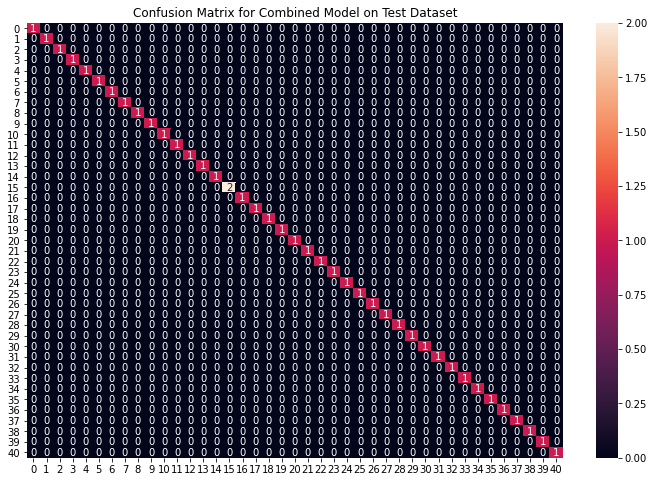
我们可以看到我们的组合模型已经准确地对所有数据点进行了分类。我们已经到了整个实现的最后一部分,我们将创建一个函数,该函数以逗号分隔的症状作为输入,并使用基于输入症状的组合模型输出预测的疾病。
创建一个可以将症状作为输入并生成疾病预测的函数
蟒蛇3
symptoms = X.columns.values
# Creating a symptom index dictionary to encode the
# input symptoms into numerical form
symptom_index = {}
for index, value in enumerate(symptoms):
symptom = " ".join([i.capitalize() for i in value.split("_")])
symptom_index[symptom] = index
data_dict = {
"symptom_index":symptom_index,
"predictions_classes":encoder.classes_
}
# Defining the Function
# Input: string containing symptoms separated by commmas
# Output: Generated predictions by models
def predictDisease(symptoms):
symptoms = symptoms.split(",")
# creating input data for the models
input_data = [0] * len(data_dict["symptom_index"])
for symptom in symptoms:
index = data_dict["symptom_index"][symptom]
input_data[index] = 1
# reshaping the input data and converting it
# into suitable format for model predictions
input_data = np.array(input_data).reshape(1,-1)
# generating individual outputs
rf_prediction = data_dict["predictions_classes"][final_rf_model.predict(input_data)[0]]
nb_prediction = data_dict["predictions_classes"][final_nb_model.predict(input_data)[0]]
svm_prediction = data_dict["predictions_classes"][final_svm_model.predict(input_data)[0]]
# making final prediction by taking mode of all predictions
final_prediction = mode([rf_prediction, nb_prediction, svm_prediction])[0][0]
predictions = {
"rf_model_prediction": rf_prediction,
"naive_bayes_prediction": nb_prediction,
"svm_model_prediction": nb_prediction,
"final_prediction":final_prediction
}
return predictions
# Testing the function
print(predictDisease("Itching,Skin Rash,Nodal Skin Eruptions,Dischromic Patches"))
输出:
{
'rf_model_prediction': 'Fungal infection',
'naive_bayes_prediction': 'Fungal infection',
'svm_model_prediction': 'Fungal infection',
'final_prediction': 'Fungal infection'
}注意:作为函数输入给出的症状在数据集中的 132 个症状中应该完全相同。