R 编程中的探索性数据分析
探索性数据分析或 EDA是一种统计方法或技术,用于分析数据集,以便通过使用一些视觉辅助工具概括其重要和主要特征。 EDA 方法可用于收集有关以下数据方面的知识:
- 数据的主要特征或特征。
- 变量及其关系。
- 找出可以在我们的问题中使用的重要变量。
EDA 是一种迭代方法,包括:
- 产生有关我们数据的问题
- 通过对我们的数据进行可视化、转换和建模来寻找答案。
- 使用我们学到的经验来改进我们的问题集或生成一组新问题。
R中的探索性数据分析
在 R 语言中,我们将在两大类下执行 EDA:
- 描述性统计,包括平均值、中位数、众数、四分位间距等。
- 图形方法,包括直方图、密度估计、箱线图等。
在我们开始使用 EDA 之前,我们必须正确执行数据检查。在我们的分析中,我们将使用 R 中的soilDB包中的loafercreek 。我们将检查我们的数据以找出所有的拼写错误和明显的错误。进一步的 EDA 可用于确定和识别异常值并执行所需的统计分析。为了执行 EDA,我们必须安装并加载以下包:
- “aqp”包
- “ggplot2”包
- “土壤数据库”包
我们可以使用install.packages()命令从 R 控制台安装这些包,并使用library()命令将它们加载到我们的 R 脚本中。我们现在将看到如何检查我们的数据并删除拼写错误和明显的错误。
R中EDA的数据检查
为了确保我们处理正确的信息,我们需要在转换过程的每个阶段清楚地查看您的数据。数据检查是在翻译之前、期间或之后查看数据以进行验证和调试的行为。现在让我们看看如何检查和删除数据中的错误和拼写错误。
例子:
R
# Data Inspection in EDA
# loading the required packages
library(aqp)
library(soilDB)
# Load from the the loakercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n < - c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p < - c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and
# add to loafercreek dataset
loafercreek$genhz < - generalize.hz(
loafercreek$hzname,
n, p)
# Extract the horizon table
h < - horizons(loafercreek)
# Examine the matching of pairing of
# the genhz label to the hznames
table(h$genhz, h$hzname)
vars < - c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
sort(unique(h$hzname))
h$hzname < - ifelse(h$hzname == "BT",
"Bt", h$hzname)R
# EDA
# Descriptive Statistics
# Measures of Central Tendency
#loading the required packages
library(aqp)
library(soilDB)
# Load from the the loakercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n <- c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p <- c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and
# add to loafercreek dataset
loafercreek$genhz <- generalize.hz(
loafercreek$hzname,
n, p)
# Extract the horizon table
h <- horizons(loafercreek)
# Examine the matching of pairing
# of the genhz label to the hznames
table(h$genhz, h$hzname)
vars <- c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
sort(unique(h$hzname))
h$hzname <- ifelse(h$hzname == "BT",
"Bt", h$hzname)
# first remove missing values
# and create a new vector
clay <- na.exclude(h$clay)
mean(clay)
median(clay)
sort(table(round(h$clay)),
decreasing = TRUE)[1]
table(h$genhz)
# append the table with
# row and column sums
addmargins(table(h$genhz,
h$texcl))
# calculate the proportions
# relative to the rows, margin = 1
# calculates for rows, margin = 2 calculates
# for columns, margin = NULL calculates
# for total observations
round(prop.table(table(h$genhz, h$texture_class),
margin = 1) * 100)
knitr::kable(addmargins(table(h$genhz, h$texcl)))
aggregate(clay ~ genhz, data = h, mean)
aggregate(clay ~ genhz, data = h, median)
aggregate(clay ~ genhz, data = h, summary)R
# EDA
# Descriptive Statistics
# Measures of Dispersion
# loading the packages
library(aqp)
library(soilDB)
# Load from the the loakercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n <- c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p <- c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and add
# to loafercreek dataset
loafercreek$genhz <- generalize.hz(
loafercreek$hzname,
n, p)
# Extract the horizon table
h <- horizons(loafercreek)
# Examine the matching of pairing of
# the genhz label to the hznames
table(h$genhz, h$hzname)
vars <- c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
sort(unique(h$hzname))
h$hzname <- ifelse(h$hzname == "BT",
"Bt", h$hzname)
# first remove missing values
# and create a new vector
clay <- na.exclude(h$clay)
var(h$clay, na.rm=TRUE)
sd(h$clay, na.rm = TRUE)
cv <- sd(clay) / mean(clay) * 100
cv
quantile(clay)
range(clay)
IQR(clay)R
# EDA
# Descriptive Statistics
# Correlation
# loading the required packages
library(aqp)
library(soilDB)
# Load from the the loakercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n <- c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p <- c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and add
# to loafercreek dataset
loafercreek$genhz <- generalize.hz(
loafercreek$hzname,
n, p)
# Extract the horizon table
h <- horizons(loafercreek)
# Examine the matching of pairing
# of the genhz label to the hznames
table(h$genhz, h$hzname)
vars <- c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
sort(unique(h$hzname))
h$hzname <- ifelse(h$hzname == "BT",
"Bt", h$hzname)
# first remove missing values
# and create a new vector
clay <- na.exclude(h$clay)
# Compute the middle horizon depth
h$hzdepm <- (h$hzdepb + h$hzdept) / 2
vars <- c("hzdepm", "clay", "sand",
"total_frags_pct", "phfield")
round(cor(h[, vars], use = "complete.obs"), 2)R
# EDA Graphical Method Distributions
# loading the required packages
library("ggplot2")
library(aqp)
library(soilDB)
# Load from the the loakercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n <- c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p <- c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and add
# to loafercreek dataset
loafercreek$genhz <- generalize.hz(
loafercreek$hzname, n, p)
# Extract the horizon table
h <- horizons(loafercreek)
# Examine the matching of pairing
# of the genhz label to the hznames
table(h$genhz, h$hzname)
vars <- c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
sort(unique(h$hzname))
h$hzname <- ifelse(h$hzname == "BT",
"Bt", h$hzname)
# graphs
# bar plot
ggplot(h, aes(x = texcl)) +geom_bar()
# histogram
ggplot(h, aes(x = clay)) +
geom_histogram(bins = nclass.Sturges(h$clay))
# density curve
ggplot(h, aes(x = clay)) + geom_density()
# box plot
ggplot(h, (aes(x = genhz, y = clay))) +
geom_boxplot()
# QQ Plot for Clay
ggplot(h, aes(sample = clay)) +
geom_qq() +
geom_qq_line()R
# EDA
# Graphical Method
# Scatter and Line plot
# loading the required packages
library("ggplot2")
library(aqp)
library(soilDB)
# Load from the the loakercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n <- c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p <- c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and add
# to loafercreek dataset
loafercreek$genhz <- generalize.hz(
loafercreek$hzname, n, p)
# Extract the horizon table
h <- horizons(loafercreek)
# Examine the matching of pairing
# of the genhz label to the hznames
table(h$genhz, h$hzname)
vars <- c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
sort(unique(h$hzname))
h$hzname <- ifelse(h$hzname == "BT",
"Bt", h$hzname)
# graph
# scatter plot
ggplot(h, aes(x = clay, y = hzdepm)) +
geom_point() +
ylim(100, 0)
# line plot
ggplot(h, aes(y = clay, x = hzdepm,
group = peiid)) +
geom_line() + coord_flip() + xlim(100, 0)输出:
> table(h$genhz, h$hzname)
2BCt 2Bt1 2Bt2 2Bt3 2Bt4 2Bt5 2CB 2CBt 2Cr 2Crt 2R A A1 A2 AB ABt Ad Ap B BA BAt BC BCt Bt Bt1 Bt2 Bt3 Bt4 Bw Bw1 Bw2 Bw3 C
A 0 0 0 0 0 0 0 0 0 0 0 97 7 7 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
BAt 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 31 8 0 0 0 0 0 0 0 0 0 0 0 0
Bt1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 2 0 0 0 0 0 0 0 8 94 89 0 0 10 2 2 1 0
Bt2 1 2 7 8 6 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 5 16 0 0 0 47 8 0 0 0 0 6
Cr 0 0 0 0 0 0 0 0 4 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
R 0 0 0 0 0 0 0 0 0 0 5 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
not-used 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0
CBt Cd Cr Cr/R Crt H1 Oi R Rt
A 0 0 0 0 0 0 0 0 0
BAt 0 0 0 0 0 0 0 0 0
Bt1 0 0 0 0 0 0 0 0 0
Bt2 6 1 0 0 0 0 0 0 0
Cr 0 0 49 0 20 0 0 0 0
R 0 0 0 1 0 0 0 41 1
not-used 0 0 0 0 0 1 24 0 0
> summary(h[, vars])
genhz clay total_frags_pct phfield effclass
A :113 Min. :10.00 Min. : 0.00 Min. :4.90 very slight: 0
BAt : 40 1st Qu.:18.00 1st Qu.: 0.00 1st Qu.:6.00 slight : 0
Bt1 :208 Median :22.00 Median : 5.00 Median :6.30 strong : 0
Bt2 :116 Mean :23.67 Mean :14.18 Mean :6.18 violent : 0
Cr : 75 3rd Qu.:28.00 3rd Qu.:20.00 3rd Qu.:6.50 none : 86
R : 48 Max. :60.00 Max. :95.00 Max. :7.00 NA's :540
not-used: 26 NA's :173 NA's :381
> sort(unique(h$hzname))
[1] "2BCt" "2Bt1" "2Bt2" "2Bt3" "2Bt4" "2Bt5" "2CB" "2CBt" "2Cr" "2Crt" "2R" "A" "A1" "A2" "AB" "ABt" "Ad" "Ap" "B"
[20] "BA" "BAt" "BC" "BCt" "Bt" "Bt1" "Bt2" "Bt3" "Bt4" "Bw" "Bw1" "Bw2" "Bw3" "C" "CBt" "Cd" "Cr" "Cr/R" "Crt"
[39] "H1" "Oi" "R" "Rt" 现在继续进行 EDA。
EDA 中的描述性统计
对于描述性统计,为了在 R 中执行 EDA,我们将所有函数分为以下几类:
- 集中趋势的度量
- 分散测量
- 相关性
我们将尝试使用集中趋势度量下的函数来确定中点值。在本节中,我们将计算均值、中位数、众数和频率。
示例 1:现在查看此示例中的集中趋势度量。
R
# EDA
# Descriptive Statistics
# Measures of Central Tendency
#loading the required packages
library(aqp)
library(soilDB)
# Load from the the loakercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n <- c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p <- c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and
# add to loafercreek dataset
loafercreek$genhz <- generalize.hz(
loafercreek$hzname,
n, p)
# Extract the horizon table
h <- horizons(loafercreek)
# Examine the matching of pairing
# of the genhz label to the hznames
table(h$genhz, h$hzname)
vars <- c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
sort(unique(h$hzname))
h$hzname <- ifelse(h$hzname == "BT",
"Bt", h$hzname)
# first remove missing values
# and create a new vector
clay <- na.exclude(h$clay)
mean(clay)
median(clay)
sort(table(round(h$clay)),
decreasing = TRUE)[1]
table(h$genhz)
# append the table with
# row and column sums
addmargins(table(h$genhz,
h$texcl))
# calculate the proportions
# relative to the rows, margin = 1
# calculates for rows, margin = 2 calculates
# for columns, margin = NULL calculates
# for total observations
round(prop.table(table(h$genhz, h$texture_class),
margin = 1) * 100)
knitr::kable(addmargins(table(h$genhz, h$texcl)))
aggregate(clay ~ genhz, data = h, mean)
aggregate(clay ~ genhz, data = h, median)
aggregate(clay ~ genhz, data = h, summary)
输出:
> mean(clay)
[1] 23.6713
> median(clay)
[1] 22
> sort(table(round(h$clay)), decreasing = TRUE)[1]
25
41
> table(h$genhz)
A BAt Bt1 Bt2 Cr R not-used
113 40 208 116 75 48 26
> addmargins(table(h$genhz, h$texcl))
cos s fs vfs lcos ls lfs lvfs cosl sl fsl vfsl l sil si scl cl sicl sc sic c Sum
A 0 0 0 0 0 0 0 0 0 6 0 0 78 27 0 0 0 0 0 0 0 111
BAt 0 0 0 0 0 0 0 0 0 1 0 0 31 4 0 0 2 1 0 0 0 39
Bt1 0 0 0 0 0 0 0 0 0 1 0 0 125 20 0 4 46 5 0 1 2 204
Bt2 0 0 0 0 0 0 0 0 0 0 0 0 28 5 0 5 52 3 0 1 16 110
Cr 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1
R 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
not-used 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 1
Sum 0 0 0 0 0 0 0 0 0 8 0 0 262 56 0 9 101 9 0 2 19 466
> round(prop.table(table(h$genhz, h$texture_class), margin = 1) * 100)
br c cb cl gr l pg scl sic sicl sil sl spm
A 0 0 0 0 0 70 0 0 0 0 24 5 0
BAt 0 0 0 5 0 79 0 0 0 3 10 3 0
Bt1 0 1 0 23 0 61 0 2 0 2 10 0 0
Bt2 0 14 1 46 2 25 1 4 1 3 4 0 0
Cr 98 2 0 0 0 0 0 0 0 0 0 0 0
R 100 0 0 0 0 0 0 0 0 0 0 0 0
not-used 0 0 0 4 0 0 0 0 0 0 0 0 96
> knitr::kable(addmargins(table(h$genhz, h$texcl)))
| | cos| s| fs| vfs| lcos| ls| lfs| lvfs| cosl| sl| fsl| vfsl| l| sil| si| scl| cl| sicl| sc| sic| c| Sum|
|:--------|---:|--:|--:|---:|----:|--:|---:|----:|----:|--:|---:|----:|---:|---:|--:|---:|---:|----:|--:|---:|--:|---:|
|A | 0| 0| 0| 0| 0| 0| 0| 0| 0| 6| 0| 0| 78| 27| 0| 0| 0| 0| 0| 0| 0| 111|
|BAt | 0| 0| 0| 0| 0| 0| 0| 0| 0| 1| 0| 0| 31| 4| 0| 0| 2| 1| 0| 0| 0| 39|
|Bt1 | 0| 0| 0| 0| 0| 0| 0| 0| 0| 1| 0| 0| 125| 20| 0| 4| 46| 5| 0| 1| 2| 204|
|Bt2 | 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 28| 5| 0| 5| 52| 3| 0| 1| 16| 110|
|Cr | 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 1| 1|
|R | 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0|
|not-used | 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 0| 1| 0| 0| 0| 0| 1|
|Sum | 0| 0| 0| 0| 0| 0| 0| 0| 0| 8| 0| 0| 262| 56| 0| 9| 101| 9| 0| 2| 19| 466|
> aggregate(clay ~ genhz, data = h, mean)
genhz clay
1 A 16.23113
2 BAt 19.53889
3 Bt1 24.14221
4 Bt2 31.35045
5 Cr 15.00000
> aggregate(clay ~ genhz, data = h, median)
genhz clay
1 A 16.0
2 BAt 19.5
3 Bt1 24.0
4 Bt2 30.0
5 Cr 15.0
> aggregate(clay ~ genhz, data = h, summary)
genhz clay.Min. clay.1st Qu. clay.Median clay.Mean clay.3rd Qu. clay.Max.
1 A 10.00000 14.00000 16.00000 16.23113 18.00000 25.00000
2 BAt 14.00000 17.00000 19.50000 19.53889 20.00000 28.00000
3 Bt1 12.00000 20.00000 24.00000 24.14221 28.00000 51.40000
4 Bt2 10.00000 26.00000 30.00000 31.35045 35.00000 60.00000
5 Cr 15.00000 15.00000 15.00000 15.00000 15.00000 15.00000现在我们将看到色散度量下的功能。在这个类别中,我们将确定中点附近的价差值。在这里,我们将计算方差、标准差、范围、四分位间距、方差系数和四分位数。
示例 2:
我们将在这个例子中看到分散的度量。
R
# EDA
# Descriptive Statistics
# Measures of Dispersion
# loading the packages
library(aqp)
library(soilDB)
# Load from the the loakercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n <- c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p <- c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and add
# to loafercreek dataset
loafercreek$genhz <- generalize.hz(
loafercreek$hzname,
n, p)
# Extract the horizon table
h <- horizons(loafercreek)
# Examine the matching of pairing of
# the genhz label to the hznames
table(h$genhz, h$hzname)
vars <- c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
sort(unique(h$hzname))
h$hzname <- ifelse(h$hzname == "BT",
"Bt", h$hzname)
# first remove missing values
# and create a new vector
clay <- na.exclude(h$clay)
var(h$clay, na.rm=TRUE)
sd(h$clay, na.rm = TRUE)
cv <- sd(clay) / mean(clay) * 100
cv
quantile(clay)
range(clay)
IQR(clay)
输出:
> var(h$clay, na.rm=TRUE)
[1] 64.89187
> sd(h$clay, na.rm = TRUE)
[1] 8.055549
> cv
[1] 34.03087
> quantile(clay)
0% 25% 50% 75% 100%
10 18 22 28 60
> range(clay)
[1] 10 60
> IQR(clay)
[1] 10现在我们将研究Correlation 。在这部分中,所有变量的所有计算相关系数值都列在表中作为相关矩阵。这为我们提供了一个量化的衡量标准,以指导我们的决策过程。
示例 3:
我们现在将在这个例子中看到相关性。
R
# EDA
# Descriptive Statistics
# Correlation
# loading the required packages
library(aqp)
library(soilDB)
# Load from the the loakercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n <- c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p <- c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and add
# to loafercreek dataset
loafercreek$genhz <- generalize.hz(
loafercreek$hzname,
n, p)
# Extract the horizon table
h <- horizons(loafercreek)
# Examine the matching of pairing
# of the genhz label to the hznames
table(h$genhz, h$hzname)
vars <- c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
sort(unique(h$hzname))
h$hzname <- ifelse(h$hzname == "BT",
"Bt", h$hzname)
# first remove missing values
# and create a new vector
clay <- na.exclude(h$clay)
# Compute the middle horizon depth
h$hzdepm <- (h$hzdepb + h$hzdept) / 2
vars <- c("hzdepm", "clay", "sand",
"total_frags_pct", "phfield")
round(cor(h[, vars], use = "complete.obs"), 2)
输出:
hzdepm clay sand total_frags_pct phfield
hzdepm 1.00 0.59 -0.08 0.50 -0.03
clay 0.59 1.00 -0.17 0.28 0.13
sand -0.08 -0.17 1.00 -0.05 0.12
total_frags_pct 0.50 0.28 -0.05 1.00 -0.16
phfield -0.03 0.13 0.12 -0.16 1.00因此,上述三个分类涉及 EDA 的描述性统计部分。现在我们将继续讨论表示 EDA 的图形方法。
EDA 中的图形方法
由于我们已经检查了我们的数据是否存在缺失值、明显错误和拼写错误,我们现在可以以图形方式检查我们的数据以执行 EDA。我们将在以下类别下看到图形表示:
- 分布
- 散点图和线图
在分布下,我们将使用条形图、直方图、密度曲线、箱线图和 QQ 图来检查我们的数据。
示例 1:
在这个例子中,我们将看到如何使用分布图来检查 EDA 中的数据。
R
# EDA Graphical Method Distributions
# loading the required packages
library("ggplot2")
library(aqp)
library(soilDB)
# Load from the the loakercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n <- c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p <- c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and add
# to loafercreek dataset
loafercreek$genhz <- generalize.hz(
loafercreek$hzname, n, p)
# Extract the horizon table
h <- horizons(loafercreek)
# Examine the matching of pairing
# of the genhz label to the hznames
table(h$genhz, h$hzname)
vars <- c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
sort(unique(h$hzname))
h$hzname <- ifelse(h$hzname == "BT",
"Bt", h$hzname)
# graphs
# bar plot
ggplot(h, aes(x = texcl)) +geom_bar()
# histogram
ggplot(h, aes(x = clay)) +
geom_histogram(bins = nclass.Sturges(h$clay))
# density curve
ggplot(h, aes(x = clay)) + geom_density()
# box plot
ggplot(h, (aes(x = genhz, y = clay))) +
geom_boxplot()
# QQ Plot for Clay
ggplot(h, aes(sample = clay)) +
geom_qq() +
geom_qq_line()
输出:
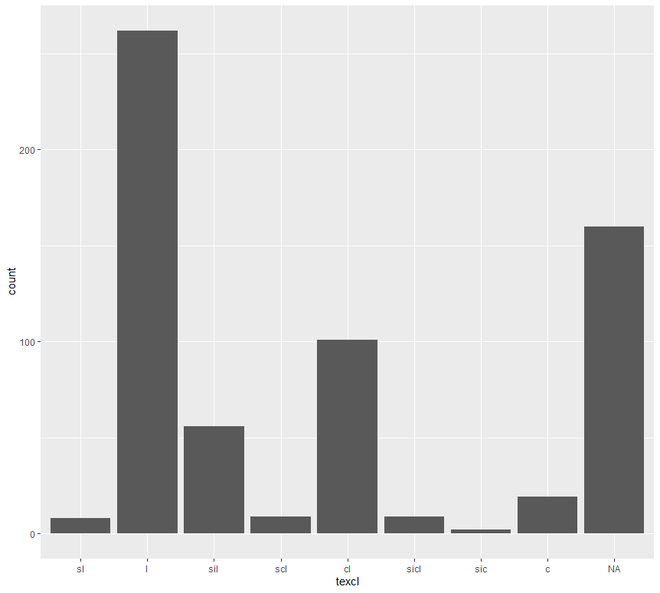

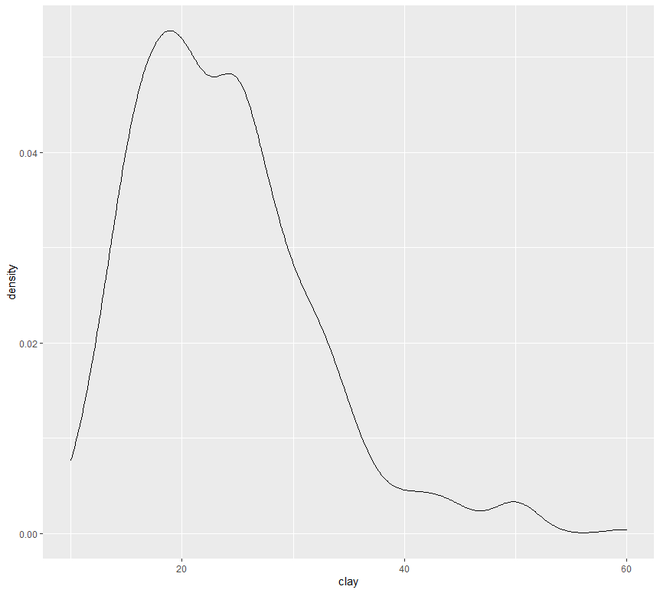
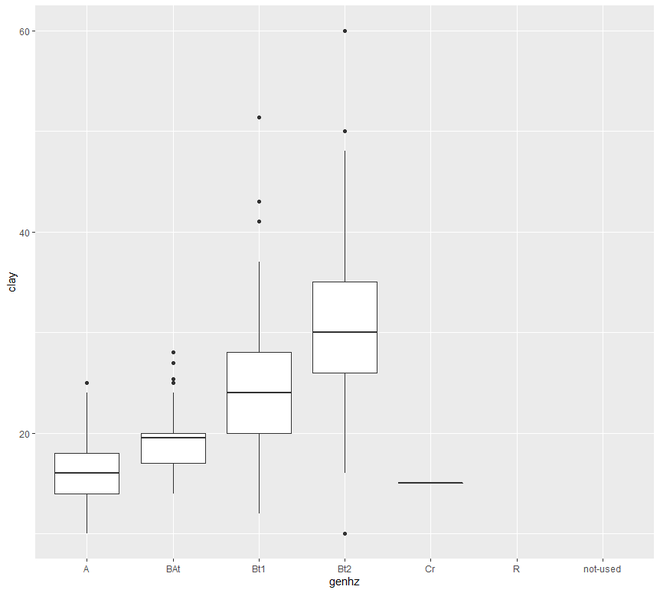
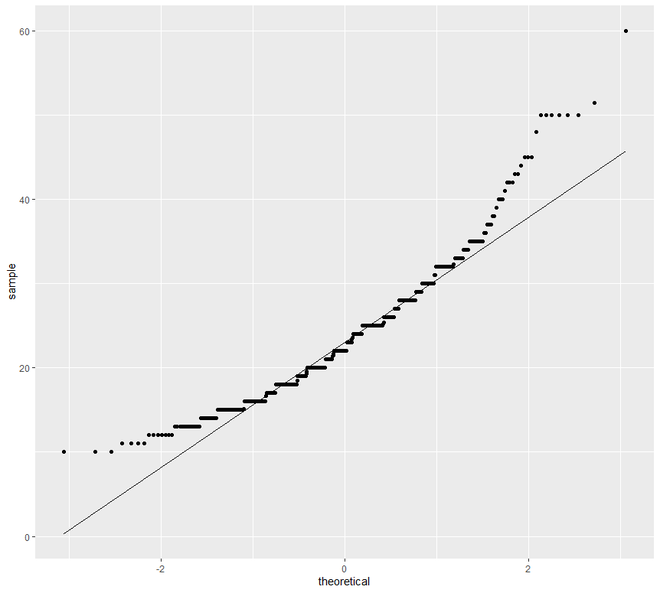
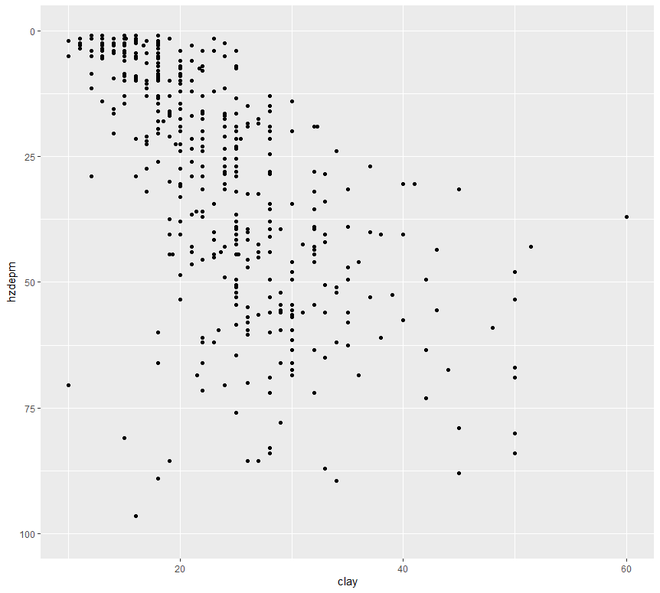
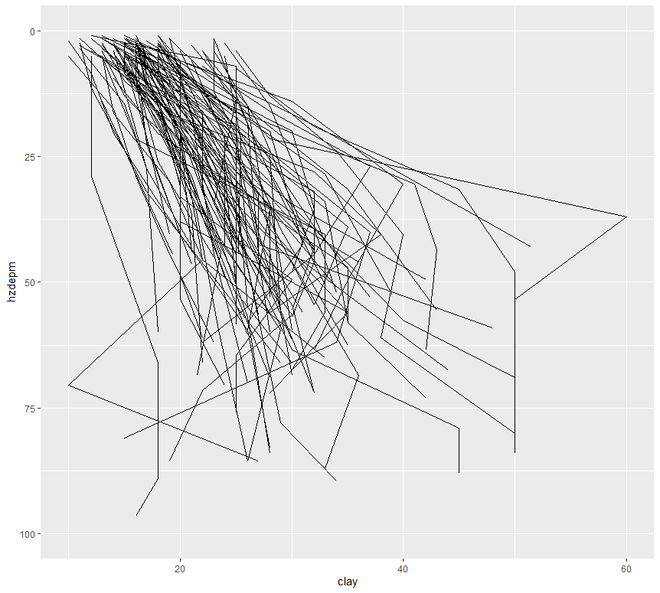
现在我们将继续进行散点图和线图。在这个类别中,我们将看到两种类型的绘图,散点图和线图。一个区间或比率变量对变量的绘图点称为散点图。
示例 2:
我们现在将了解如何使用散点图和折线图来检查我们的数据。
R
# EDA
# Graphical Method
# Scatter and Line plot
# loading the required packages
library("ggplot2")
library(aqp)
library(soilDB)
# Load from the the loakercreek dataset
data("loafercreek")
# Construct generalized horizon designations
n <- c("A", "BAt", "Bt1", "Bt2", "Cr", "R")
# REGEX rules
p <- c("A", "BA|AB", "Bt|Bw", "Bt3|Bt4|2B|C",
"Cr", "R")
# Compute genhz labels and add
# to loafercreek dataset
loafercreek$genhz <- generalize.hz(
loafercreek$hzname, n, p)
# Extract the horizon table
h <- horizons(loafercreek)
# Examine the matching of pairing
# of the genhz label to the hznames
table(h$genhz, h$hzname)
vars <- c("genhz", "clay", "total_frags_pct",
"phfield", "effclass")
summary(h[, vars])
sort(unique(h$hzname))
h$hzname <- ifelse(h$hzname == "BT",
"Bt", h$hzname)
# graph
# scatter plot
ggplot(h, aes(x = clay, y = hzdepm)) +
geom_point() +
ylim(100, 0)
# line plot
ggplot(h, aes(y = clay, x = hzdepm,
group = peiid)) +
geom_line() + coord_flip() + xlim(100, 0)
输出: