Biopython – 成对对齐
成对序列比对是一次比较两个序列并提供最佳可能序列比对的过程。成对序列比对使用动态规划算法。 Biopython 有一个特殊的模块Bio.pairwise2 ,它使用成对方法识别比对序列。与其他软件相比,Biopython 提供了寻找比对序列的最佳算法。
让我们以两个简单的假设序列为例来使用成对模块。
Python3
# Import libraries
from Bio import pairwise2
from Bio.Seq import Seq
# Creating sample sequences
seq1 = Seq("TGTGACTA")
seq2 = Seq("CATGGTCA")
# Finding similarities
alignments = pairwise2.align.globalxx(seq1, seq2)
# Showing results
for match in alignments:
print(match)Python3
# Import libraries
from Bio import pairwise2
from Bio.Seq import Seq
from Bio.pairwise2 import format_alignment
# Creating sqmple sequences
seq1 = Seq("TGTGACTA")
seq2 = Seq("CATGGTCA")
# Finding similarities
alignments = pairwise2.align.globalxx(seq1, seq2)
# Showing results
for alignment in alignments:
print(format_alignment(*alignment))Python3
# Import libraries
from Bio import Align
from Bio.Seq import Seq
# Creating sample sequences
seq1 = Seq("TGTGACTA")
seq2 = Seq("CATGGTCA")
# Calling method
aligner = Align.PairwiseAligner()
# Showing method attributes
print(aligner)
# Finding similarities
alignments = aligner.align(seq1, seq2)
# Showing results
for alignment in alignments:
print(alignment)输出:
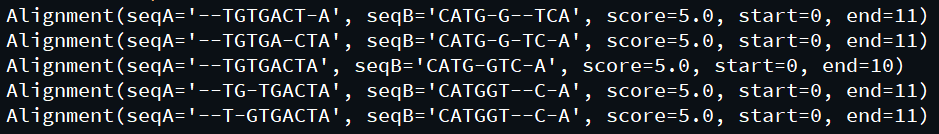
在这里, globalxx方法完成主要工作,它遵循约定
匹配参数:Code Character Description x No parameters. Identical character has score of 1, else 0. m match score of identical chars, else mismatch score. d dictionary returning scores of any pair of characters. c A callback function returns scores.
差距惩罚参数:Code Character Description x No gap penalties. s both sequences having same open and extend gap penalty. d sequences having different open and extend gap penalty. c A callback function returns the gap penalties.
对于一个漂亮的打印输出Bio.pairwise2提供了format_alignment()方法:
蟒蛇3
# Import libraries
from Bio import pairwise2
from Bio.Seq import Seq
from Bio.pairwise2 import format_alignment
# Creating sqmple sequences
seq1 = Seq("TGTGACTA")
seq2 = Seq("CATGGTCA")
# Finding similarities
alignments = pairwise2.align.globalxx(seq1, seq2)
# Showing results
for alignment in alignments:
print(format_alignment(*alignment))
输出:
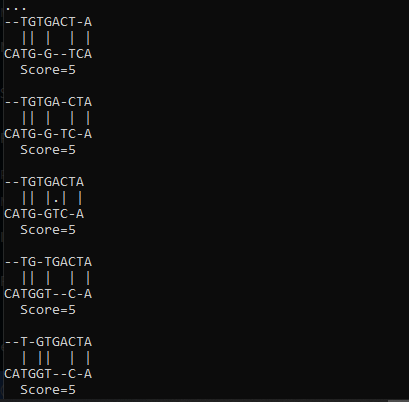
Biopython提供了另一个模块来进行成对序列比对。为此, Align模块有一个PairwiseAligner() 。它有各种 API 来设置参数,如模式、匹配分数、算法、差距惩罚等。 下面是该方法的简单实现:
蟒蛇3
# Import libraries
from Bio import Align
from Bio.Seq import Seq
# Creating sample sequences
seq1 = Seq("TGTGACTA")
seq2 = Seq("CATGGTCA")
# Calling method
aligner = Align.PairwiseAligner()
# Showing method attributes
print(aligner)
# Finding similarities
alignments = aligner.align(seq1, seq2)
# Showing results
for alignment in alignments:
print(alignment)
输出:
