二次判别分析
线性判别分析
现在,让我们考虑由贝叶斯概率分布 P(Y=k | X=x) 表示的分类问题,LDA 通过尝试对给定预测变量类别(即 Y 的值)P(X =x| Y=k):
![]()
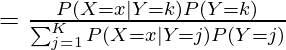
在 LDA 中,我们假设 P(X | Y=k) 可以估计为多元正态分布,该分布由以下等式给出:
![]()
在哪里, 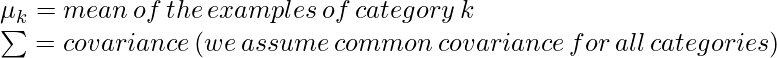
和 P(Y=k) =\pi_k。现在,我们尝试用以下假设写出上面的等式:
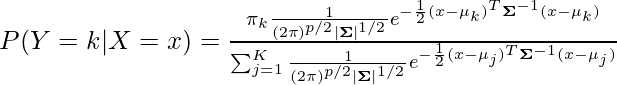
现在,我们对两边取对数并最大化方程,我们得到决策边界:
![]()
对于两个类,决策边界是 x 的线性函数,其中两个类的值相等,该线性函数为:
![]()
对于多类 (K>2),我们需要估计 pK 均值、pK 方差、K 先验比例和![]() .现在,我们更详细地讨论二次判别分析。
.现在,我们更详细地讨论二次判别分析。
二次判别分析
二次判别分析与线性判别分析非常相似,只是我们放宽了所有类的均值和协方差相等的假设。因此,我们需要单独计算。
现在,对于每个 y 类,协方差矩阵由下式给出:
通过添加以下项并求解(同时取对数 和 )。二次判别函数由下式给出:
![]()
执行
- 在这个实现中,我们将使用 R 和 MASS 库来绘制线性判别分析和二次判别分析的决策边界。为此,我们将使用 iris 数据集:
R
# import libraries
library(caret)
library(MASS)
library(tidyverse)
# Code to plot decision plot
decision_boundary = function(model, data,vars, resolution = 200,...) {
class='Species'
labels_var = data[,class]
k = length(unique(labels_var))
# For sepals
if (vars == 'sepal'){
data = data %>% select(Sepal.Length, Sepal.Width)
}
else{
data = data %>% select(Petal.Length, Petal.Width)
}
# plot with color labels
int_labels = as.integer(labels_var)
plot(data, col = int_labels+1L, pch = int_labels+1L, ...)
# make grid
r = sapply(data, range, na.rm = TRUE)
xs = seq(r[1,1], r[2,1], length.out = resolution)
ys = seq(r[1,2], r[2,2], length.out = resolution)
dfs = cbind(rep(xs, each=resolution), rep(ys, time = resolution))
colnames(dfs) = colnames(r)
dfs = as.data.frame(dfs)
p = predict(model, dfs, type ='class' )
p = as.factor(p$class)
points(dfs, col = as.integer(p)+1L, pch = ".")
mats = matrix(as.integer(p), nrow = resolution, byrow = TRUE)
contour(xs, ys, mats, add = TRUE, lwd = 2, levels = (1:(k-1))+.5)
invisible(mats)
}
par(mfrow=c(2,2))
# run the linear disciminant analysis and plot the decision boundary with Sepals variable
model = lda(Species ~ Sepal.Length + Sepal.Width, data=iris)
lda_sepals = decision_boundary(model, iris, vars= 'sepal' , main = "LDA_Sepals")
# run the quadratic disciminant analysis and plot the decision boundary with Sepals variable
model_qda = qda(Species ~ Sepal.Length + Sepal.Width, data=iris)
qda_sepals = decision_boundary(model_qda, iris, vars= 'sepal', main = "QDA_Sepals")
# run the linear disciminant analysis and plot the decision boundary with Petals variable
model = lda(Species ~ Petal.Length + Petal.Width, data=iris)
lda_petal =decision_boundary(model, iris, vars='petal', main = "LDA_petals")
# run the quadratic disciminant analysis and plot the decision boundary with Petals variable
model_qda = qda(Species ~ Petal.Length + Petal.Width, data=iris)
qda_petal =decision_boundary(model_qda, iris, vars='petal', main = "QDA_petals")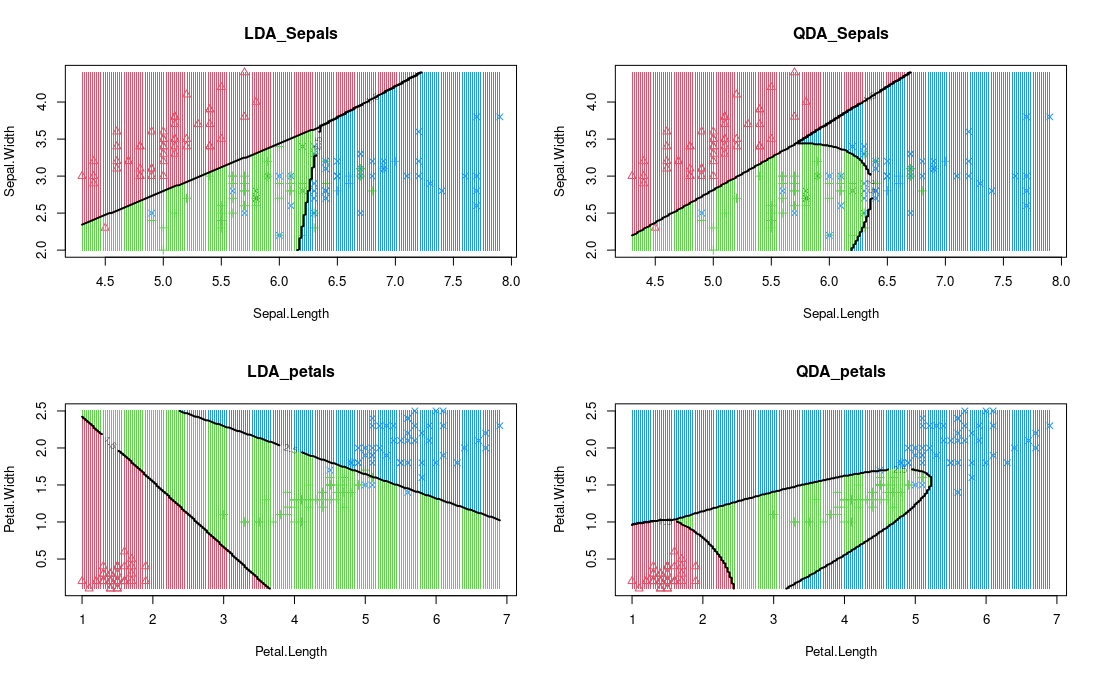
LDA 和 QDA 可视化
参考:
- 斯坦福统计笔记
- 哈佛关于 LDA 的幻灯片